
DE
|
EN
Quality control
Maldi
Maldi-TOF mass spectroscopy
|
Before shipment biomers.net checks the integrity and correctness of each individual oligonucleotide (if applicable). With Maldi-TOF mass spectrometry (matrix assisted laser desorption/ionisation; time of flight) the molecular mass enables a definite statement about composition and integrity of an oligonucleotide.
The principle of this method is such, that a single molecule will need a defined time to cover a given distance (time of flight), depending on the exact molecular mass. By using appropriate support material, oligos can be excited by laser to "fly" in an ionised state across a vacuum to the detector. The time needed for this distance is the basis to calculate the molecular mass.
|
 |
|
With Maldi, differences as low as one single hydrogen atom can be resolved. When working with oligonucleotides, however, different orders of magnitude are of interest. A complete nucleotide for example (e.g. when looking for n-1-products) has a molecular mass around 300. If present in detectable amount, n-1-products will thus be clearly identified.
Also, the successful coupling of modifications can be easily controlled by Maldi; most flourescent dyes e.g. have molecular masses in the range of 500- 800 Dalton.
Only an oligo comprising the complete sequence (and the desired modifications, where applicable) will show in the Maldi spectroscopy the calculated molecular mass!
|
|
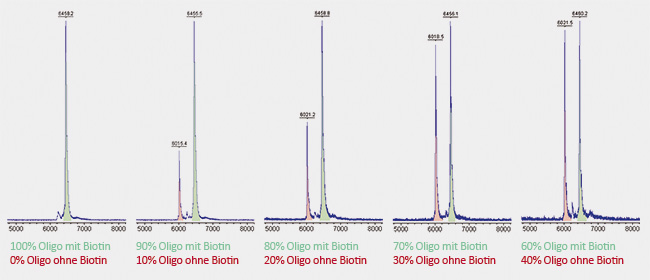
Here, two identical oligonucleotides were synthesized, each 20 bases long. One of them was coupled to biotin at the
5´-terminus, the other remained without biotin. Both oligonucleotides were dissolved in the same molarity and mixed in different ratios with each other. A significant peak can already be seen at admixture of only 10% of unmodified oligo (shown in red). Due to its relatively long linker, biotin has a molecular weight of about 430 Da. Accordingly to this, the red peak of the unmodified oligo is shifted by 430 Da to the left compared to the green peak of the biotin-carrying oligo. Due to its lower weight, the unmodified oligo requires less time to reach the detector. |
 |
|
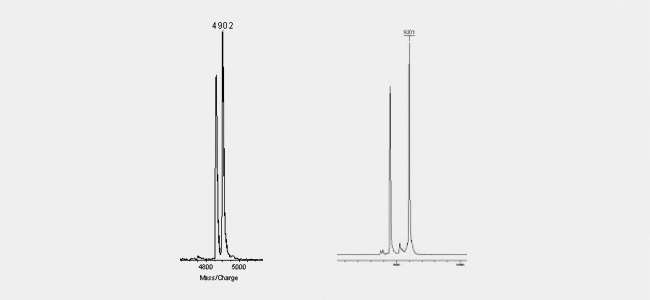
Wobble bases (figure left) are as clearly detectable as decay-products know for example from BlackHole quenchers (right figure). Theses quenchers are know to break due to the laser beam into defined products. One part will be kept attached to the oligo, the other will fall off. As this decay is generally not complete, but will affect just a fraction of the oligos, the Maldi shows two individual peaks: one comprising the correct molecular mass as calculated from the sequence and the coupled modifications and an additional peak with the calulated molecular mass minus the lost part of the quencher.
|
 |
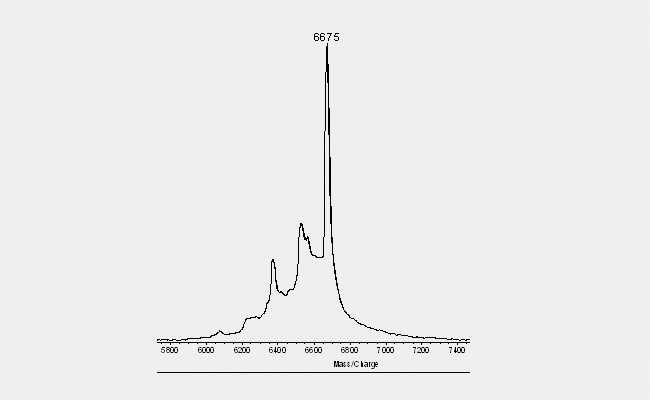
| Another typical picture (see below) obtain with Maldi spectrograms derives from purins within the sequence. Depending on the exact composition of the complete oligonucleotide, distinct purins tend to "fall off" when hit by the laser during the Maldi measurement. This event gives the spectrogramm the appearance of a "ladder" with decreasing peak height to the left. The peaks are separated by 150 Dalton, the molecular mass of a purin. As this occurs only due to the laser beam and the Maldi is performed only with an aliquot of the final product, such a picture gives no indication about a loss of quality in the final product. |
 |
ESI
Electrospray Ionisation
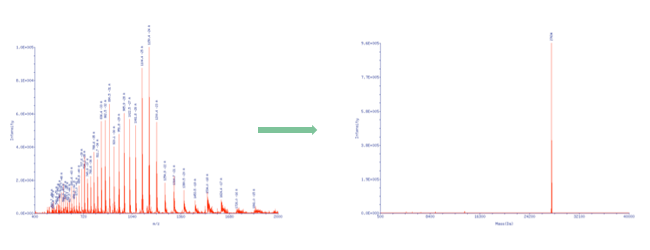
| By using ESI mass spectroscopy it is possible to determine the mass of oligonucleotides in solution by direct injection into the ion source or alternatively by means of a LC unit equipped with a reversed phase column. The mild ionization from solution makes ESI-MS a very good choice especially for longer oligonucleotides. When compared to Maldi spectra the raw data doesn’t show a single molecular ion peak but a series of signals due to multiply charged cations in positive mode and anions in negative mode. In the case of oligonucleotides multiply charged anions are formed when spraying the slightly alkaline analyte solution in the ion source and thus a signal pattern, which allows to calculate the exact molecular mass by means of a mathematical transformation. |
|
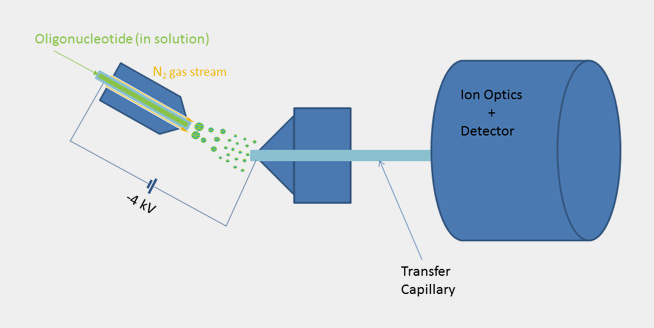 |
CGE
Capillary gel electrophoresis
| High sensitivity and resolution of CGE is based on the combination of electrophoresis and a gel sieving by molecular size together with a miniaturized capillary kept at constant temperature. A capillary filled with polymer solution is more suitable for high throughput CE than cross-linked gel, though, because it can be replaced easily between runs for identical separating conditions. Only a small amount of analyte is needed for injection into the polymer filled capillary. After high voltage is applied, the oligonucleotides in the mixture separate in the electric field because of their difference in mobility due to different molecular size and, depending on modifications, net charge. Number and amount of by-products including the n-1 oligonucleotide is then quantified by UV detection. |
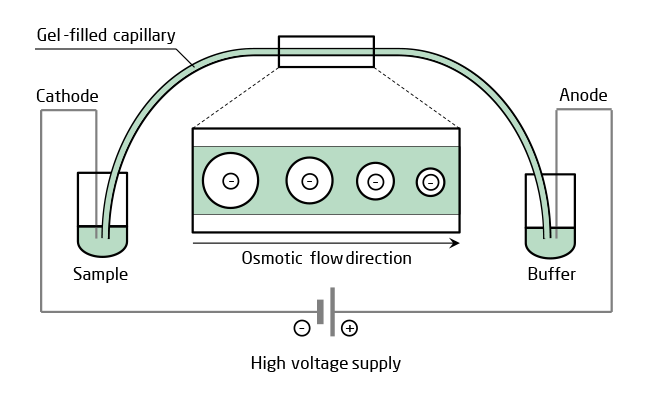 |

