
Real-Time PCR Probes
By real time PCR a technique is understood, which is observable during its performance by using appropriate tools . In contrast to a normal PCR, where the terminal result is analysed on an agarose-gel, in a real-time PCR the increase of the DNA amount can be observed and detected directly - in real time.
The direct observation of each individual cycle is the basis for quantification, which has opended a broad field of new applications particularly in diagnostics and validation.
The direct observation is attained by using flourescent dyes. This can be achieved by SybrGreen® or related products, attaching to double stranded DNA and hence indicating the generation of new double strands in each cycle.
More elegant and highly sensitive, but more sophisticated and not always applicable is the use of probes. Probes are added to the PCR reaction in addition to the primers and will bind to the amplified region, increasing thus already the specificity. Using probes with appropriate fluorophore/quencher combination enables a direct detection of the binding.
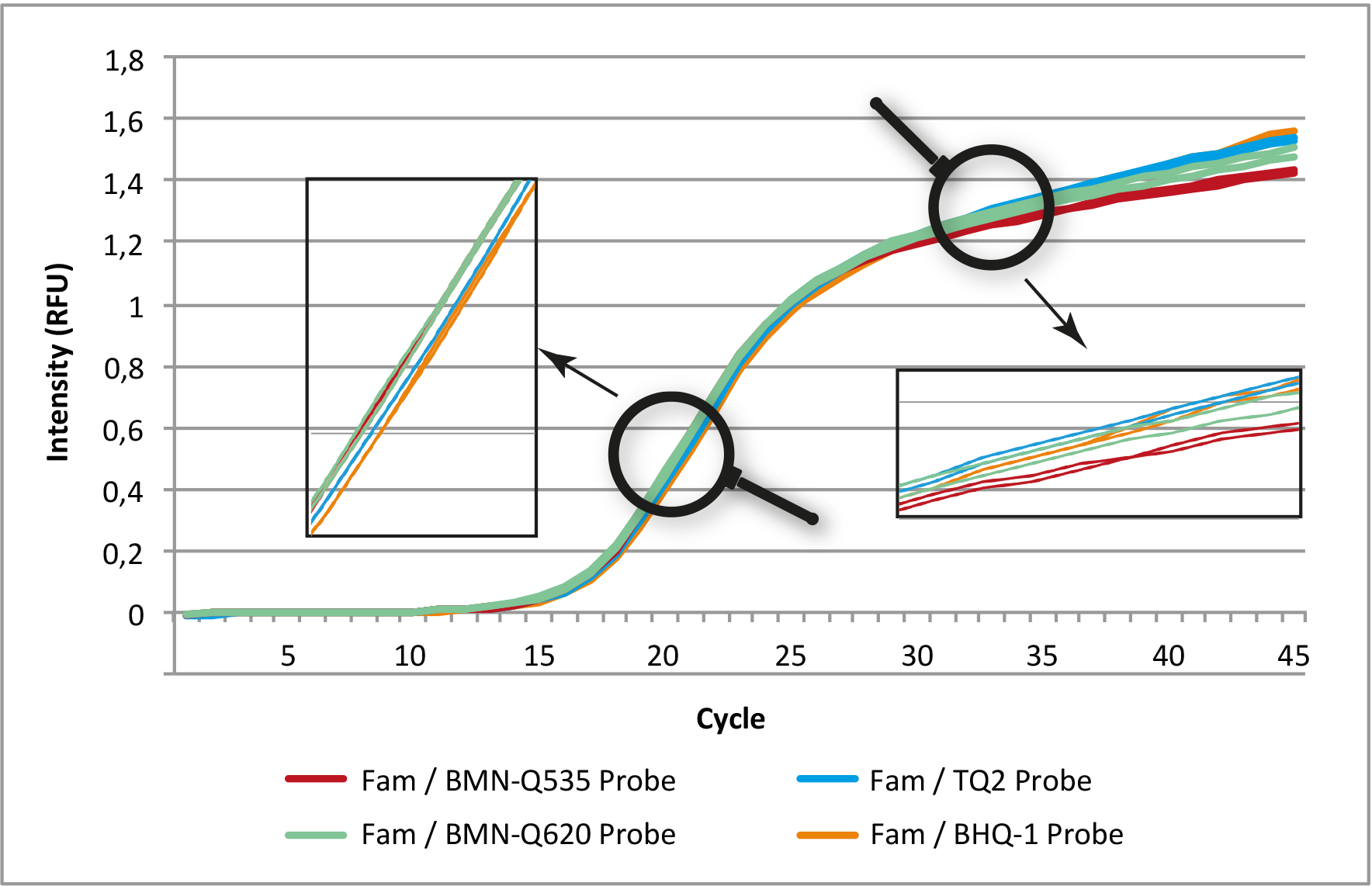
Figure 1: Comparison of amplification curves (normalized) of a qPCR assay with 5'-6-Fam-labeled probes and four different 3'-quenchers.
In each assay, the same primer and probe sequences were used. All assays were run in duplicate (Genaxxon ProbeMasterMix No ROX,
MyGo Pro, pre-calibrated channel for Fam).
In direct comparison, the quenchers (BMN-Q535, BMN-Q620, BHQ-1 and TQ2) show almost identical results with the fluorescence dye 6-Fam.
Basically, five 'types' of experimental set-ups are used, which are described here shortly.
DL-Probes - BMN-Q
Dual-Labelled Probes with BMN-Q460, BMN-Q535, BMN-Q620, BMN-Q1 and BMN-Q2
In the so-called TaqMan®-Assay a dual labelled probe is placed on the target such that it comes to sit "in the way" of the polymerase.
In the free and in the hybridised state the proximity of fluorophore and quencher is close enough to quench the fluorescence. During the PCR, however, the 3'-5' -exonuclease activity of the polymerase will digest the probe, freeing the fluorophore from the influence of the quencher.
On request, we offer a design-service for dual labelled probes to use in TaqMan Assays.
Dual-Labelled Probes with BMN quenchers
| 5'-Fluorophor | ABS [nm] | EM [nm] | recommended BMN 3´-Quencher |
|---|---|---|---|
| Atto 425 | 436 | 484 | BMN-Q1, BMN-Q535, BMN-Q460 |
| Atto 465 | 453 | 508 | BMN-Q1, BMN-Q535, BMN-Q460 |
| Fam | 494 | 520 | BMN-Q1, BMN-Q535 |
| Joe | 520 | 548 | BMN-Q1, BMN-Q535 |
| Hex | 535 | 556 | BMN-Q1, BMN-Q535 |
| Cyanine 3 | 550 | 570 | BMN-Q1, BMN-Q535 |
| Atto 550 | 554 | 579 | BMN-Q1, BMN-Q535 |
| Atto Rho 11 | 572 | 595 | BMN-Q2, BMN-Q620 |
| Rox | 574 | 602 | BMN-Q2, BMN-Q620 |
| Cyanine 3.5 | 588 | 604 | BMN-Q2, BMN-Q620 |
| Atto Rho 101 | 586 | 610 | BMN-Q2, BMN-Q620 |
| Atto 647N | 646 | 664 | BMN-Q2, BMN-Q620 |
| Cyanine 5 | 649 | 670 | BMN-Q2, BMN-Q620 |
| Cyanine 5.5 | 675 | 694 | BMN-Q2, BMN-Q620 |
* All prices are valid for DNA oligonucleotides (max. 30 bases) with the respective dye-quencher-combinations; incl. HPLC-purification and Maldi-quality control. The price is valid for online orders (biomers.net online order form). VAT, shipping costs and duties may apply. Additional conditions are according to our current price list. Prices may change without further notice. Errors excluded.
DL-Probes - BHQ, TQ, BBQ, Tamra
Dual-Labeled probes with BHQ, BBQ, TQ and Tamra quenchers
| 5'-Fluorophor | ABS [nm] | EM [nm] | 3'-Quencher |
|---|---|---|---|
| Atto 425 | 436 | 484 | BHQ-1 BBQ-650 |
| Fam | 494 | 520 | TQ2 BHQ-1 BBQ-650 Tamra |
| Joe | 520 | 548 | TQ2 BHQ-1 |
| Hex | 535 | 556 | TQ2 BHQ-1 BBQ-650 |
| Cyanine 3 | 550 | 570 | TQ3 BBQ-650 BHQ-2 |
| Atto 550 | 554 | 579 | BHQ-2 BBQ-650 |
| Atto Rho 11 | 572 | 595 | BHQ-2 |
| Rox | 574 | 602 | TQ3 BBQ-650 BHQ-2 |
| Cyanine 3.5 | 588 | 604 | TQ3 BBQ-650 BHQ-2 |
| Atto Rho 101 | 586 | 610 | BHQ-2 |
| Atto 647N | 646 | 664 | BHQ-2 BBQ-650 |
| Cyanine 5 | 649 | 670 | BBQ-650 BHQ-2 |
| Cyanine 5.5 | 675 | 694 | BBQ-650 |
* All prices are valid for DNA oligonucleotides (max. 30 bases) with the respective dye-quencher-combinations; incl. HPLC-purification and Maldi-quality control. VAT, shipping costs and duties may apply. Additional conditions are according to our current price list. Prices may change without further notice. Errors excluded.
DQ-Probes
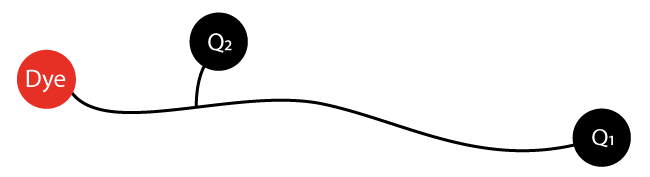
Increased qPCR sensitivity with Double Quenched Probes from biomers.net
Standard 5'-nuclease assay qPCR probes have a fluorophor at the 5'-end and an appropriate quencher at the 3'-end of the oligonucleotide. Besides quenching the emitted fluorescence of the 5'-dye, the quencher molecule also prevents elongation of the probe oligonucleotide during PCR.
For longer probes with increasing distance between fluorophor and quencher one may observe an increase of unwanted background. In these cases incorporation of an additional quencher (e.g. BMN-Q535 or BMN-Q620) into the oligonucleotide sequence near the 5'-fluorophor highly improves the qPCR sensitivity and lowers the background.
Double Quenched Probes consist of a 5'-fluorophor, a 3'-BMN quencher and an additional internal BMN quencher which is in a distance of 8 - 10 bases from the fluorophor.
| Dual-Labelled Probe | Double Quenched Probe |
|---|---|
|
|
 |
| 5'-Fam, Hex, Cyanine, Atto-Dyes, etc. 3'-BMN-Q535, BMN-Q620 |
5'-Fam, Hex, Rox, Cyanine, Atto-Dyes, etc. int. BMN-Q535, BMN-Q620 3'-BMN-Q535, BMN-Q620 |
Standard Dual Labelled Probes as well as Double Quenched Probes with BMN quenchers are available for online ordering; for Double Quenched Probes please have a look at the ordering information.
A detailed ordering information you can find here.
For any further information or help, please contact the biomers.net customer service.
MGB
Minor Groove Binder (MGB)
MGB-modified oligonucleotides show increased melting temperature (Tm) and specificity and can form very stable duplexes with complementary DNA sequences. Here, the base composition of the oligos strongly influences the melting temperature of the modified oligos.
When binding the probe to the target sequence, minor groove binders (MGB) bind to the minor groove of the DNA. The crescent shape of the MGB CDPI3 (Dihydropyrroloindolecarboxylate Tripeptide CDPI3) allows an exact spatial adaptation to the curved structure of the minor groove, thus allowing an intercalation between the sugar-phosphate chains of the DNA backbone. This isohelical conformation is stabilised mainly by van der Waals forces and hydrophobic interactions.
According to this, an extremely high stability of the double-stranded molecule can be achieved, so that even very short DNA probes can hybridise very efficiently to the target DNA.
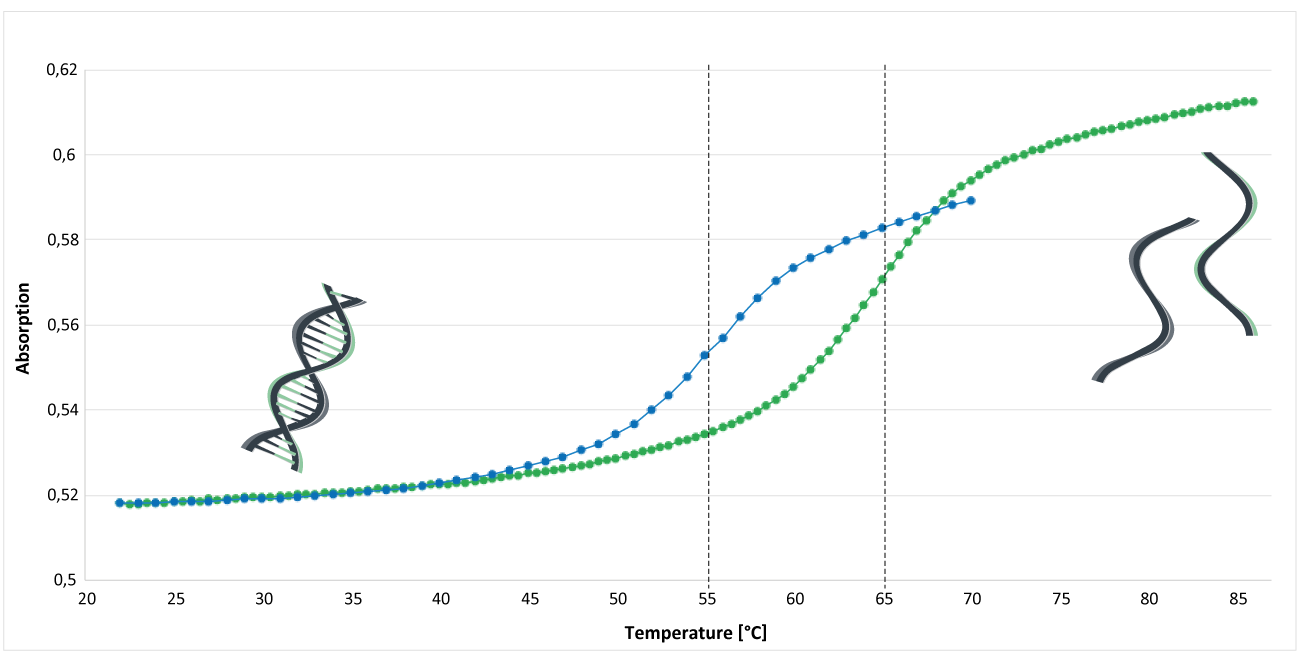
Figure 1 shows the analysis of the melting curve of an MGB CDPI3-modified and an unmodified oligo. The analysis was performed in phosphate buffer (pH 7.4). The concentration of Na+ was 30 mM; the concentration of the DNA single strands was approx. 1.3 μM. The inflection point of the melting curve indicates the melting temperature Tm of the oligos.
The blue curve represents an unmodified EUB338 oligo: 5'-gctgcctcccgtaggagt-3 '. The melting temperature of this oligo is around 55°C.
The green curve shows the melting curve of an MGB CDPI3-modified oligo. The EUB oligo was labelled at the 3 'end with a MGB CDPI3 modification: 5'-gctgcctcccgtaggagt-MGB CDPI3. Here, the melting temperature is around 65°C.
Under these conditions, comparing these two oligos, an increase in melting temperature of about 10°C can be detected.
A detailed ordering information you can find here.
For any further information or help, please contact the biomers.net customer service.
Literature:
1. Minor Groove Binder-Conjugated DNA Probes for Quantitative DNA Detection by Hybridization-Triggered Fluorescence. Afonina IA, ReedMW, Lusby E, Shishkina IG, Belousov YS; BioTechniques (2002) 32:940-949 .
2. 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J; Nucleic Acids Research (2000), Volume 28, Issue 2, Pages 655–661.
Beacons
Molecular Beacons
Another variant is given by Molecular Beacons®. Similar to Scorpions® probes, which have been developed on the basis of Molecular Beacons, these molecules are designed to contain a 'stem' structure, a short region of complementary sequence at both ends. This enables the molecule to fold back with itself, bringing the terminally coupled fluorophore and quencher in close proximity. The specific probe sequence, which is complementary to the target sequence, forms the so-called ´loop´ structure in this conformation.
In the absence of a suitable target molecule the terminal coupled modifications (dye and quencher) are held in close proximity by the ´stem´, so that the fluorescence is inhibited. In this state, the fluorophore transfers its electrons to the quencher, which in turn emits this energy either as light of a longer wavelength or as heat.
In the presence of a suitable target molecule, the beacon opens by binding of the 'loop' region to the corresponding target sequence. This probe-target hybrid is longer and designed to be more stable than binding of the 'stem' region. This finally leads to the opening of the molecular beacon. The conformational change separates the fluorophore and the quencher to release the fluorescence.
Molecular beacons are highly specific and in the best of cases, are capable of identifying differences based on a single nucleotide. The length of the probe sequence is selected to form a perfect complementary probe-target hybrid molecule which is just more stable than the complementary 'stem' region. Already a difference of one nucleotide in the probe sequence reduces stability of the probe-target molecule significantly, so that the stability of the'stem' region is predominant and opening of the molecular beacon is prevented.
Due to their special features Molecular Beacons are very versatile:
- PCR
- Detection of specific RNAs in living cells1
- Amplicon detection in dignostic assays
- Multiplex assays to detect different targets in one reaction
- SNP detection
- Pharmacogenetic applications
- Genetic screening (determination of genotype at a particular locus)
www.molecular-beacons.org provides elaborated information on molecular beacons and their applications.
We are pleased to offer a design service for Molecular Beacons®, Scorpions® and TaqMan® probes.
Just send us the sequence of the target you want to analyse. Our experienced specialists will design your individual probe.
If you would like more information about this topic, please contact us at any time.
Tel +49 731 70 396 0 ǀ info@biomers.net
| 5‘-Fluorophor | 3‘-Quencher |
|---|---|
| 6-Fam, Hex, Tet | BMN-Q535, Dabcyl, DDQ-l, BHQ-1, TQ-2, BBQ-650 |
| Yakima Yellow | BHQ-1, TQ-2, BBQ-650 |
| Cyanine 3, Cyanine 5, Tamra, Rox, Texas Red |
BMN-Q620, BHQ-2, TQ-3, BBQ-650 |
| Atto dye, DY dye | BMN-Q, BHQ, TQ, BBQ-650 |
Prices can be found in our current price list. Further combinations on request.
Literature:
1. Real time detection of DNA.RNA hybridization in living cells. Sokol DL, Zhang X, Lu P, Gewirtz AM; Proc Natl Acad Sci USA (1998),11538-43.
Scorpions
Scorpions
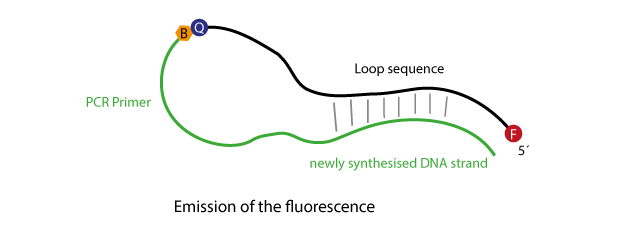
Scorpions primer are multiple modified oligonucleotides which are used in a particular variant of Real time PCR to generate highly specific signals.
Scorpions-oligos bear at their 5´-end a fluorophore, which in the unhybridised state is kept in close proximity to a quencher by a particularly designed stem structure. Further 3´, following the quencher, a target specific primer is attached. This in combination with an unmodified 'reverse' pimer is used in a PCR reaction to amplify the target region. To prevent the polymerase from filling up the complete Scorpions-sequence, a further modification is included, a so-called blocker, leading to a stop of polymerisation near the quencher, thus, the loop/stem-region of the Scorpions-oligo will remain as single-strand.
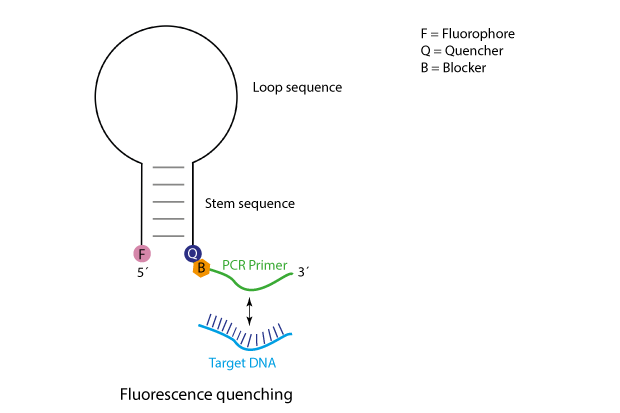
In a Scorpions PCR the basic events are as follows:
Initially the primer sequence of the Scorpions oligo will anneal to its respective target sequence. After the primer has been elongated, the stem structure will open and compete with the binding of the newly synthesised strand.
The opening of the stem enables the fluorophore to escape the influence of the quencher, thus, the fluorescence now can be used for detection.
biomers.net offers Scorpions at the following conditions:
| 5‘-Fluorophor | Quencher | Blocker |
|---|---|---|
| 6-Fam, Hex, Tet | int. BHQ-1, int. Dabcyl | Spacer18 |
| Cyanine 3, Cyanine 5 | int. BHQ-2, int. Dabcyl | Spacer18 |
| Atto dye, DY dye | int. BHQ | Spacer18 |
Prices can be found in our current price list. Further combinations on request.
Scorpions are produced with a licence from DxS.
Prices are inclusive DNA-oligonucleotide up to 60 bases, 5´-fluorophore, internal coupling of the quencher as well as the polymerase blocker, HPLC-purification and Maldi quality control and are valid only for electronic ordering.
Fret Probes
Fret Probes
A related application is the LightCycler®-Assay. Here two probes are placed beside each other on the target. This alone already increases the sensitivity of the assay significantly, enabling differentiation of closely related genes or organisms.
The binding of both probes is detected using a FRET-transfer (FRET: fluorecsence resonance energy transfer). While exciting the fluorophore on the "left" side, its emission is not detected directly, but used to excite the second probe, which in turn now emitts light at its appropriate wavelength.


