
Modifications for various applications
Gadolinium-DTPA
Gadolinium-DTPA (Gd-DTPA) modified oligonucleotides
Gadolinium ions (Gd) have strong paramagnetic properties and may influence the spin relaxation of neighbouring protons. This effect is exploited in biochemical NMR studies, or even more important, in magnetic resonance imaging (MRI) in medical field, where Gd complexes are used as contrast agents.
Under certain circumstances, oligonucleotide aptamers (DNA or RNA oligonucleotides) can accumulate in specific target tissues. If these aptamers are conjugated with Gd ions, this can lead to higher-contrast images in corresponding imaging procedures.
The binding of gadolinium ions to oligonucleotides usually occurs through the corresponding chelate complexes (e.g. Gd-DTPA, Gd-DOTA). Generally, the oligonucleotide is first conjugated with a chelating agent. In the next step, the metal ions (in this case Gd) are “captured” then and efficiently bound to form an extremely stable complex.
For the labelling of oligonucleotides, various chelating agents and different coupling strategies are available. The picture below exemplifies gadolinium-DTPA modification at the 5´-end of an oligonucleotide.
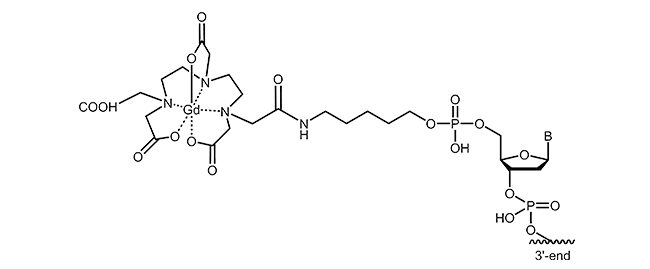
Literature:
1. Gadolinium(III) Complexes with N-Alkyl-N-methylglucamine Surfactants Incorporated into Liposomes as Potential MRI Contrast Agents. Silva RS, Correia Duarte E, Ramos GS, Kock FVC, Diuk Andrade F, Frézard F, Colnago LA, Demicheli C; Bioinorganic Chemistry and Applications (2015), Volume 2015, Article ID 942147, 8 pages.
2. Characteristics of gadolinium-DTPA complex: a potential NMR contrast agent. Weinmann HJ, Brasch RC, Press WR, Wesbey GE. AJR Am J Roentgenol. (1984); 142(3):619-24.
3. Gadolinium-DTPA as a contrast agent in MRI: initial clinical experience in 20 patients. Carr DH, Brown J, Bydder GM, Steiner RE, Weinmann HJ, Speck U, Hall AS, Young IR; American Journal of Roentgenology. (1984); 143: 215-224. 10.2214/ajr.143.2.215.
4. Molecular MR Contrast Agents for the Detection of Cancer: Past and Present. Bogdanov A, Mazzanti ML; Semin Oncol. (2011); 38(1): 42–54.
5. Macromolecular MRI contrast agents: Structures, properties and applications. Tang J, Sheng Y, Hu H, Shen Y; Progress in Polymer Science (2013); Volume 38, Issues 3–4, Pages 462-502.
6. Gadolinium conjugated TiO2-DNA oligonucleotide nanoconjugates show prolonged intracellular retention period and T1-weighted contrast enhancement in Magnetic Resonance images. Paunesku T, Ke T, Dharmakumar R, Mascheri N, Wu A, Lai B, Vogt S, Maser J, Thurn K, Szolc-Kowalska B, Larson A, Bergan RC, Omary R, Li D, Lu Z-R, Woloschak GE; Nanomedicine (2008); 4(3): 201–207.
Puromycin
Puromycin
|
Puromycin is one of the aminonucleoside antibiotics which can be isolated from streptomyces strain Streptomyces alboniger. Puromycin shows structural similarity with the base adenosine or N6-N6-dimethyladenosine. Its effect is based on the inhibition of protein biosynthesis by binding to the 50S subunit of the ribosome via a peptide bond and thus affecting the growth of prokaryotic and eukaryotic cells.1 As an analogue of an aminoacyl-tRNA, puromycin can bind to the ribosomal A-site and integrates into the growing polypeptide chain. This leads to immediate termination of chain elongation, the ribosomal subunit dissociates and finally the translation stops.2 After termination, puromycin is located at the C-terminus of the truncated, newly synthesized protein and inhibits continuation of synthesis.3 |
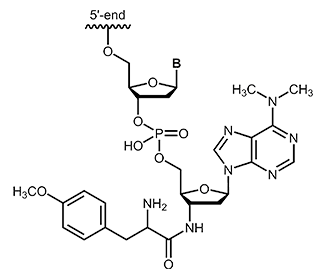 |
biomers.net offers puromycin as 3´-modification with a maximum length of 45 bases.
Literature:
1. Molecular cloning and characterization of gene encoding novel puromycin-inactivating enzyme from blasticidin S-producing Streptomyces morookaensis. Nishimura M, Ikeda K, Sugiyama M; J Biosci Bioeng. (2006), 101(1):63-9.
2. Identification and characterization of a drug-sensitive strain enables puromycin-based translational assays in Saccharomyces cerevisiae. Cary GA, Yoon SH, Torres CG, Wang K, Hays M, Ludlow C, Goodlett DR, Dudley AM; Yeast (2014), (5):167-78.
3. Puromycin oligonucleotides reveal steric restrictions for ribosome entry and multiple modes of translation inhibition. Starck SR, Roberts RW; RNA (2002), 8:890–903.
Acridinium ester
Acridinium ester
In molecular biology, acridinium esters are commonly used for selective labeling of proteins and nucleic acids1. The acridinium ester can be bound to the corresponding target molecule (DNA, protein) by a covalent linkage2. According to their highly sensitive chemiluminescence releasing in the presence of hydrogen peroxide, acridinium esters also can be used for clinical diagnostics. In contrast to radiolabeled DNA, acridinium ester linkages provide a sensitive, stable and above all safe detection method in immunoassays3.
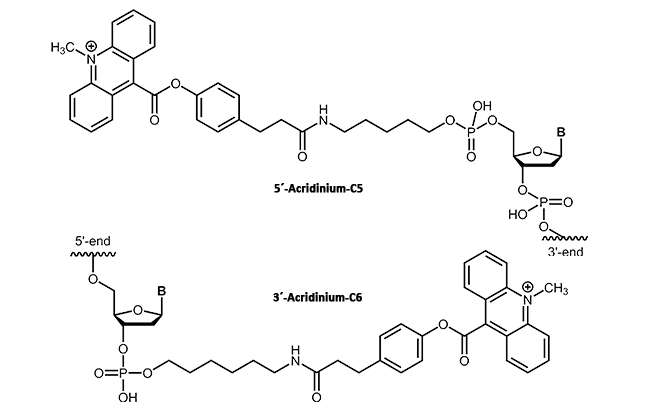 |
biomers.net offers acridinium ester as 3'- or 5'-modification up to a length of 45 bases.
Literature:
1. Acridinium ester chemiluminescence: pH dependent hydrolysis of reagents and flow injection analysis of hydrogen peroxide and glutamate. Stuever Kaltenbach M, Arnold MA; Microchimica Acta (1992), Volume 108, Issue 3-6, pp 205-219.
2. Acridinium ester-labelled DNA oligonucleotide probes. Septak M; J Biolumin Chemilumin. (1989), (1):351-6.
3. Acridinium esters as high-specific-activity labels in immunoassay. Weeks I, Beheshti I, McCapra F, Campbell AK, Woodhead JS; Clin Chem. (1983), (8):1474-9.
4-Carboxymethylaniline
Modifications for the immobilisation on surfaces
Carboxymethylaniline (4-CMA) allows to immobilise coupled biomolecules to appropriate prepared surfaces. In this way, the binding of oligonucleotides to conductive surfaces (e.g. graphite) enables the detection of currents. By coupling, either to the 5 'or 3' end of oligonucleotides, the molecules are covalently bonded in the desired orientation to the surface.
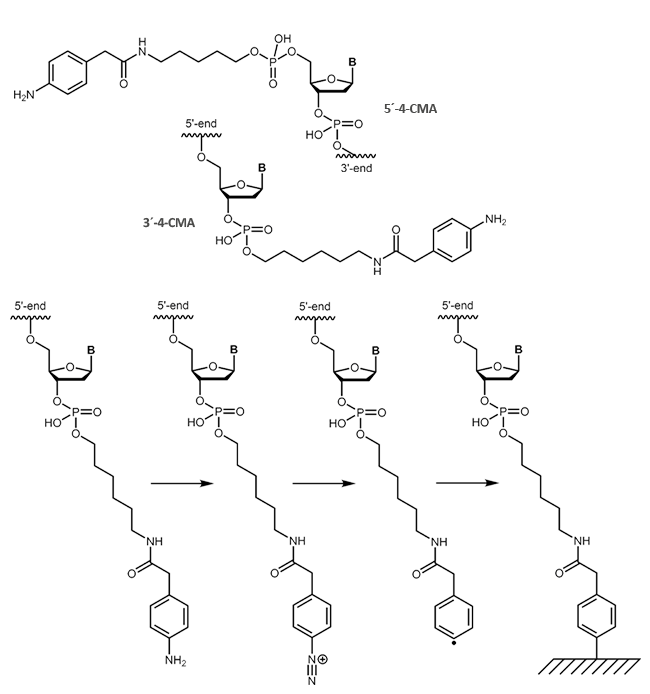 |
Literature:
- Diazonium-Protein Adducts for Graphite Electrode Microarrays Modification: Direct and Addressed Electrochemical Immobilization. Corgier BP, Marquette CA, Blum LJ; Journal of the American Chemical Society, (2005), 127, 18328-18332.
- A versatile method for direct and covalent immobilisation of DNA and proteins on biochips. Corgier BP, Laurent A, Perriat P, Blum LJ, Marquette CA; Angewandte Chemie International (2007), 46, 4108-4110.
- On-Chip Chemiluminescent Signal Enhancement using Nanostructured Gold-Modified Carbon Microarrays. Corgier BP, Li F, Blum LJ, Marquette CA; Langmuir (2007), 23(16), 8619-8623.
Methacrylamide
Immobilisation on surfaces via Methacrylamide-modified oligonucleotides
Methacrylamide-modified, also known as acrydite-modified oligonucleotides are an important part and parcel of molecular biology research for many years. By polymerising of free acrylic acid monomers (formation of polyacrylamides) or by coupling with thiol, methacrylamide-modified oligonucleotides can be covalently bound to surfaces. By means of this fast and simple attachment to surfaces, desired DNA single strands can be immobilised, enriched, identified or purified.
A well-known example is the biochip technology (microarrays) in which the methacralymide-modified oligonucleotides are bound to a thiol-labelled glass surface and thus accessible to single-stranded DNA.
This results in a variety of possible applications:
- Monitoring of mRNA expression
- Sequencing of DNA
- Genotyping
- Identification of SNP
- Detection of viruses, bacteria and other pathogens
biomers.net offers 5'- and 3'-methacrylamide-modified DNA oligonucleotides.
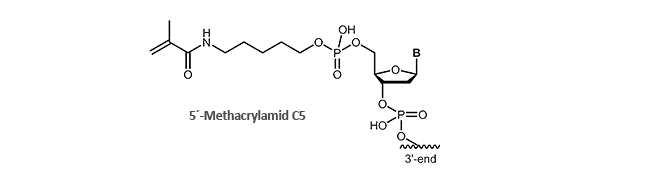
Literature:
1. Immobilization of acrylamide-modified oligonucleotides by co-polymerization. Rehman FN, Audeh M, Abrams ES, Hammond PW, Kenney M, Boles TC; N. Acids Research. 27, (1998), Bd. 2, 649-655.
2. Mutation typing using electrophoresis and gel-immobilized Acrydite probes. Kenney M, Ray S, Boles TC; Biotechniques (1998), (3):516-21.
Gemcitabine
Oligonucleotides with the cytostatic drug gemcitabine dFdC
Gemcitabine (2´,2´-difluoro deoxycytidine, dFdC) is an analogue of the pyrimidine nucleoside deoxycytidine (dC). Gemcitabine differs from dC in two fluorine atoms (instead of two hydrogen atoms) at the 2´-position of the sugar.
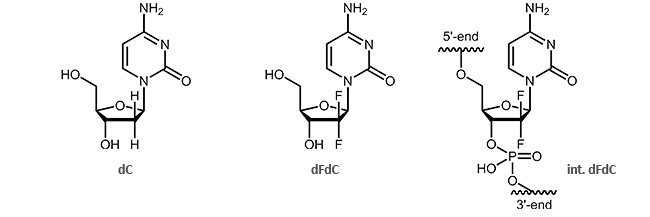
The effect of the two additional fluorine atoms is shown in the inhibition of DNA synthesis.1,2 It is particularly interesting that after incorporation of gemcitabine, DNA can only be extended by one further nucleotide. Then, the synthesis interrupts and is blocked thus resulting the death of the cell. This process is called “masked chain termination” since the last nucleotide prevents the cytidine analogue dFdC from detection and degradation by exonucleases or DNA repair enzymes.3 Due to its activity as cytostatic drug, gemcitabine is preferably used in chemotherapy as an anti-tumor agent.3
biomers.net now offers dFdC for incorporation into oligonucleotides!
The internal coupling is possible at any desired position of the oligo. Also multiple couplings can be synthesised. Combinations with other modifications are possible on request.
Literature:
1. 2',2'-Difluoro-deoxycytidine (gemcitabine) incorporation into RNA and DNA of tumour cell lines. Ruiz van Haperen VW, Veerman G, Vermorken JB, Peters GJ; Biochem Pharmacol. (1993); 46(4):762-6.
2. Quantification of gemcitabine incorporation into human DNA by LC/MS/MS as a surrogate measure for target engagement. Wickremsinhe ER, Lutzke BS, Jones BR, Schultz GA, Freeman AB, Pratt SE, Bones AM, Ackermann BL; Anal Chem. (2010);82(15):6576-83. doi: 10.1021/ac100984h.
3. DNA Repair in Cancer Therapy: Molecular Targets and Clinical Applications. Kelley MR; Academic Press (2012), 95-98.
4. Synthesis and restriction enzyme analysis of oligodeoxyribonucleotides containing the anti-cancer drug 2',2'-difluoro-2'-deoxycytidine. Richardson FC, Richardson KK, Kroin JS, Hertel LW; Nucleic Acids Res. (1992); 20(7): 1763–1768.
CoA
Oligonucleotide-Coenzyme A conjugates
Many fields of research require a specific linking of native proteins with building blocks, affecting the transport behavior or allow specific immobilization. Using small 'tags' which can be fused to the proteins, proteins can be combined almost arbitrarily without compromising their folding or function. A highly flexible system makes use of the selective linking of the so-called ybbR tags with coenzyme A.
We are pleased to be able to offer coenzyme A-modified oligos. This opens up many possibilities of DNA-protein chimeras.
An impressive example can be found at:
Protein–DNA Chimeras for Nano Assembly (Pippig et al., ACS Nano, 2014)
Ask...we like to discuss your individual project!
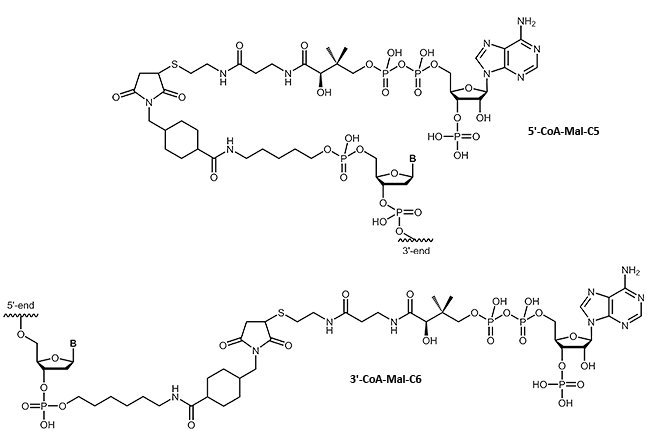 |
Literature:
- Protein-DNA Chimeras for Nano Assembly. Pippig DA, Baumann F, Strackharn M, Aschenbrenner D, Gaub HE; ACS Nano (2014), 8 (7), pp 6551–6555.
- The Ribosome Modulates Nascent Protein Folding. Kaiser CM, Goldman DH, Chodera JD, Tinoco Jr. I, Bustamante C; Science (2011), 334(6063): 1723–1727. doi:10.1126/science.1209740.

