Antisense Oligos
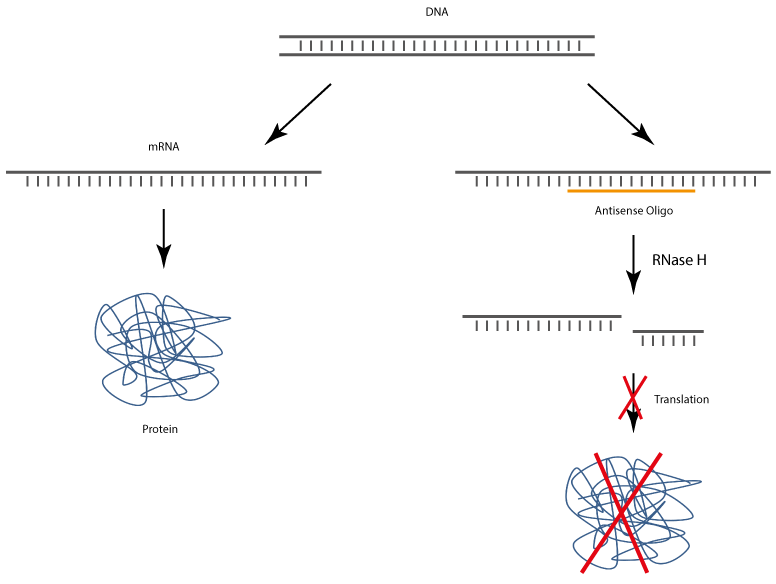
After uptake into target cells antisense oligonucleotides bind to corresponding complementary mRNA, whereby the translation and also the biosynthesis of the target protein is specifically inhibited.
The complex of antisense DNA oligonucleotide and the bound RNA strand is recognised by RNase H, which enzymatically cleaves the RNA. As a result, the antisense DNA oligonucleotide is released and thus again available for binding complementary RNA.
Some critical aspects are the cellular uptake, as the negatively charged DNA can only poorly pass through cell membranes. Here, various lipophilic molecules have proven to be very helpful.
Furthermore, unprotected, short DNA oligonucleotides are rapidly degraded in cells. In order to prevent or at least slow down the degradation, in addition to phosphothioates various 2´-modifications are successfully used.
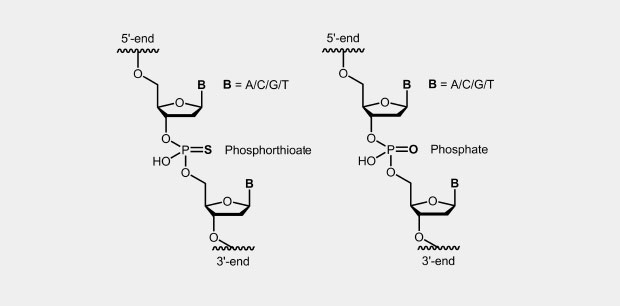
Phosphorothioate
Phosphorothioate Oligonucleotides (Phosphorothioate, PTO)
The phosphorothioate modification of the DNA backbone is one of the first and still widely used modification for antisense oligonucleotides (antisense oligos of the first generation). In the phosphate backbone, one of the two oxygen atoms which is not involved in the internucleotide bridge, is replaced by a sulfur atom (Fig.). Due to the four different substituents, the central phosphorus atom is a chiral center which effects diastereomer formation.
In oligonucleotides, all (Full-PTO) or just single phosphates (Part-PTO) can be replaced by phosphorothioate. Often it seems to be sufficient to incorporate two to four PTOs at the end of the oligos to achieve the desired stability against degradation.
|
|
Order information:
Phosphorothioates are presented in the order system of biomers.net by asterisks (*). If all or just single phosphodiester should be replaced by phosphorothioates, a * have to be inserted into the sequence at the respective position.
Examples:
- A normal sequence without phosphorothioates:
agcgtgacgtggacgtgagtga - The identical sequence, but as a "full-PTO-oligo" (all inter-nucleotidic phosphodiester were changed to PTO):
a * g * c * t * g * g * a * c * t * g * g * g * a * c * t * g * g * a * g * t * g * a - The sequence with single PTOs, in this case three PTO at both ends:
a * g * c * gtgacgtggacgtgag * t * g * a - The sequence with a PTO-bridge at the 3 'end:
agcgtgacgtggacgtgagtg * a
If a PTO-Oligo should be modified at one end, for example with a fluorescent dye, it is often useful, to "stabilize" also the phosphate bridge between the oligo and the modification (synthesizing as a PTO).
At a 5'-modification, in this case a * is also added to the 5'-end of the oligos (at a 3'-modification according to the 3'-end) and the desired modification is selected in the menu:
- * a * g * c * t * g * g * a * c * t * g * g * g * a * c * t * g * g * a * g * t * g * a, 5'-modification: biotin: PTO between biotin and oligo
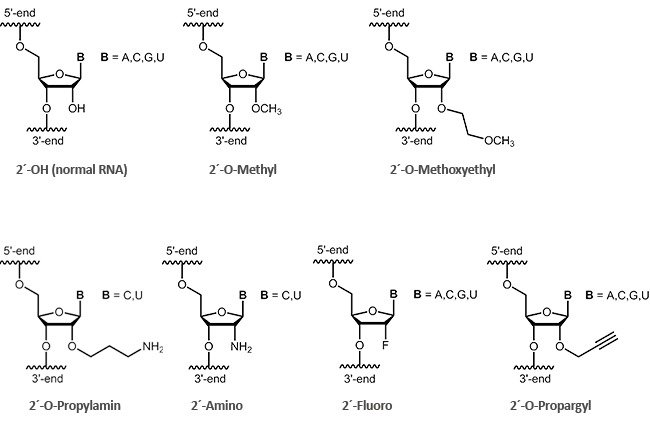
2´-modified RNA
2´-modified RNA
Antisense oligos of the second generation include RNA oligonucleotides with alkyl modifications at the 2´-position of the ribose sugar. The two most important representatives of these modifications are 2´-O-methyl (OMe) and 2´-O-methoxyethyl (MOE) RNAs.
By definition it is a ribose if an oxygen is linked at 2´-carbon of the sugar. Antisense oligonucleotides having a modification at the 2'-hydroxyl group (2´-O-methyl, 2´-O-methoxyethyl), often show an increased stability to enzymatic degradation. This also facilitates an improved distribution of the oligos in the cell.1 Furthermore, it was observed that 2´-modified oligonucleotides (2´-O-methyl, 2´-O-methoxyethyl, 2´-O-propyl, 2´-O-fluoro) bind with greater affinity to complementary RNA.2
biomers.net offers here a selection of different modifications:
|
 |
Please contact the biomers.net customer support for availability and delivery times.
Tel +49 731 70 396 0 | info@biomers.net
Literature:
1. Pharmacokinetic Properties of 2´-O-(2-Methoxyethyl)-Modified Oligonucleotide Analogs in Rats. Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, Sioufi NB, Samor H, Manoharan M, Levin AA; The Journal of Pharmacology and experimental Therapeutics (2001), Vol. 296, No. 3; JPET 296:890–897.
2. Evaluation of 2´-Modified Oligonucleotides Containing 2’-Deoxy Gaps as Antisense Inhibitors of Gene Expression. Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM; The Journal of Biological Chemistry (1993), Inc. Val. 268, No. 19, pp. 14514-14522.
Gapmer Oligos
Gapmer oligonucleotides
Generally, antisense oligos are able to regulate gene expression by inhibiting the translation mechanism and inducing the enzymatic cleavage of the target RNA.
The antisense oligonucleotides of the first or second generation (PTO- or 2´-modified oligos) frequently used in the past have an increased stability in the serum in contrast to unmodified oligos, but show toxic side effects in comparison to other antisense oligonucleotides which can greatly limit the range of applications of these oligonucleotides.
PTO-modified antisense oligonucleotides also show lower binding capacities to complementary bases (reduced Tm) and non-specific binding to proteins.
Newer generation antisense oligos are so-called gapmers. A gapmer is a single-stranded DNA oligo that can specifically bind to endogenous target mRNA sequences. The resulting mRNA gapmer duplex is recognised by
RNase H, which induces the enzymatic degradation of the mRNA and thus inhibits the specific expression of the target gene.
Type H ribonucleases are ubiquitous enzymes that are found both in the cell nucleus and in the cytoplasm. RNase H is a sequence-unspecific endonuclease that hydrolyses the RNA of RNA-DNA heteroduplexes after an antisense oligo has been specifically bound to the target mRNA. In this way, the degraded mRNA cannot be translated into the corresponding protein, which leads to down-regulation of the target protein (gene silencing).
Gapmer structure
Gapmers are chimeric oligonucleotides consisting of a central block of DNA nucleotides flanked by modified bases on both sides. Here, 2´-O-modified ribonucleotides as well as other artificially modified nucleotide monomers (e.g. LNA) are possible, which protect the DNA block against enzymatic nuclease degradation.
Usually, seven to eight unmodified DNA bases are required as a central block in order to fully activate RNase H and to initiate cleavage of the target mRNA. At the 3´-and 5´-ends it has been shown that three modified bases are sufficient to protect the oligo against exonuclease degradation and to increase the half-life of the oligo tenfold.
Such optimally designed gapmer oligos achieve a higher affinity and sequence specificity for the complementary counter strand and show a high biological stability with at the same time low toxicity in biological serum and cell medium. In LNA gapmers, for example, with each inserted LNA base the melting temperature can be increased by 1.5-4°C and a higher affinity for complementary sequences can be achieved.
Literature:
- Design of antisense oligonucleotides stabilized by locked nucleic acid. Kurreck J, Wyszko E, Gillen C, Erdmann VA; Nucleic Acids Research (2002), Vol. 30, No.9.
- GapmeR cellular internalization by macropinocytosis induces sequence-specific gene silencing in human primary T-cells. Fazil MHUT, Ong ST, Chalasani MLS, Low JH, Kizhakeyil A, Mamidi A, Lim CFH, Wright GD, Lakshminarayanan R, Kelleher D, Verma NK. Scientific Reports (2016), 6:37721.
Chimera
Chimeric oligonucleotides
Chimeric oligonucleotides can be made from various types of building blocks. In contrast to naturally occurring molecules, synthetic oligos can simultaneously consist of DNA and RNA or PNA. It is also possible that these oligos are mixed from right- and left-handed components (see L-DNA, L-RNA, PNA). Furthermore, mixtures are possible with reverse building blocks, whereby molecules can be generated that terminate at both ends with 3 'or 5'.
Please contact us for any further information.
Literature:
- An Antisense Oligonucleotide Primer. Hogrefe RI; Antisense and Nucleic Acid Drug Development (1999), 9(4): 351-357.
Modifications
Antisense Oligonucleotide Modifications
An ineffectual uptake of oligonucleotide conjugates to the cell often represent the greatest difficulty for a successful development of new oligonucleotide-based therapeutics.
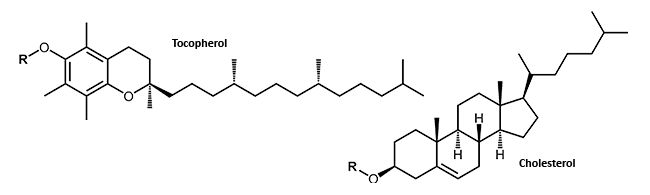
In terms of antisense oligonucleotides, a wide variety of modifications are available. Matched to the researcher´s particular method, various internal or terminal changes of an oligonucleotide are possible. Small lipophilic molecules which can be attached to an oligonucleotide may facilitate the passage of the highly negatively charged DNA or RNA through the cell membrane and thus the uptake into the cell.
Small lipophilic molecules which can be attached to oligonucleotide, allow the passage through the cell membrane and thus the uptake into the cell of the highly negatively charged DNA or RNA. The attachment of cholesterol or tocopherol to DNA or RNA molecules may improve the stability and the distribution of the conjugates in the cell1 (Lipophilic Modifications).
 |
|
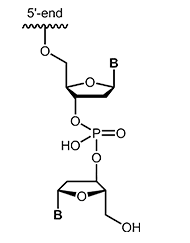
Modifications which may also inhibit the degradation of the oligos by nucleases are inverted-ends. A further nucleotide is attached "upside down" to the 3'-terminal nucleotide so that the oligonucleotide has a 5'-terminus on both sides. Besides protection against digestion, this modification efficiently blocks elongation during PCR. |
 |
biomers.net offers in terms of antisense oligos to a wide range of ligands and further modifications. Our experienced support team will help you any time at all aspects of the selection and use of your specific oligonucleotides.
Literature:
1. Oligonucleotide conjugates for therapeutic applications. Winkler J; Ther. Deliv. (2013), 4(7),791-809.
Literature:
- An antisense oligonucleotide primer. Hogrefe RI; Antisense and Nucleic Acid Drug Development (1999), 9(4): 351-357.
- Prospects for nucleic acid-based therapeutics against hepatitis C virus. Lee CH, Kim JH, Lee S-W. World J Gastroenterol. (2013), 19(47): 8949–8962.
- CpG-A and CpG-B oligonucleotides differentially enhance human peptide–specific primary and memory CD8 T-cell responses in vitro. Rothenfusser S, Hornung V, Ayyoub M, Britsch S, Towarowski A, Krug A, Sarris A, Lubenow N, Speiser D, Endres S, Hartmann G; Blood (2004), Vol.103, Number 6.
- CpG-A Oligonucleotides Induce a Monocyte-Derived Dendritic Cell-Like Phenotype That Preferentially Activates CD8 T Cells. Krug A, Rothenfusser S, Selinger S, Bock C, Kerkmann M, Battiany J, Sarris A, Giese T, Speiser D, Endres S, Hartmann G;. J Immunol (2003), 170:3468-3477.