Hapten modifications
Haptens are small molecules that can be recognized by specific antibodies and, bound to an oligonucleotide, allow detection, localization or isolation with antibody-based methods. Haptenes can serve as non-radioactive labels on oligonucleotide probes in Southern, Northern and in situ hybridisation (ISH) followed by immuno-detection or for purification methods based on affinity chromatography. The biotin-streptavidin system, that takes advantage of the specific and very strong biotin-streptavidin interaction, makes an analogue approach.
| Abbreviation | Modification | ||||
|---|---|---|---|---|---|
| 5’ | 3’ | 5’ / 3’ dual | internal | ||
| Biotin | Bio | x | x | x | x |
| Digoxigenin | DIG | x | x | x | x |
| Dinitrophenol | DNP | x | x | x | |
| Fluorescein | 6-Fam FITC |
x x |
x x |
x x |
x x |
Biotin
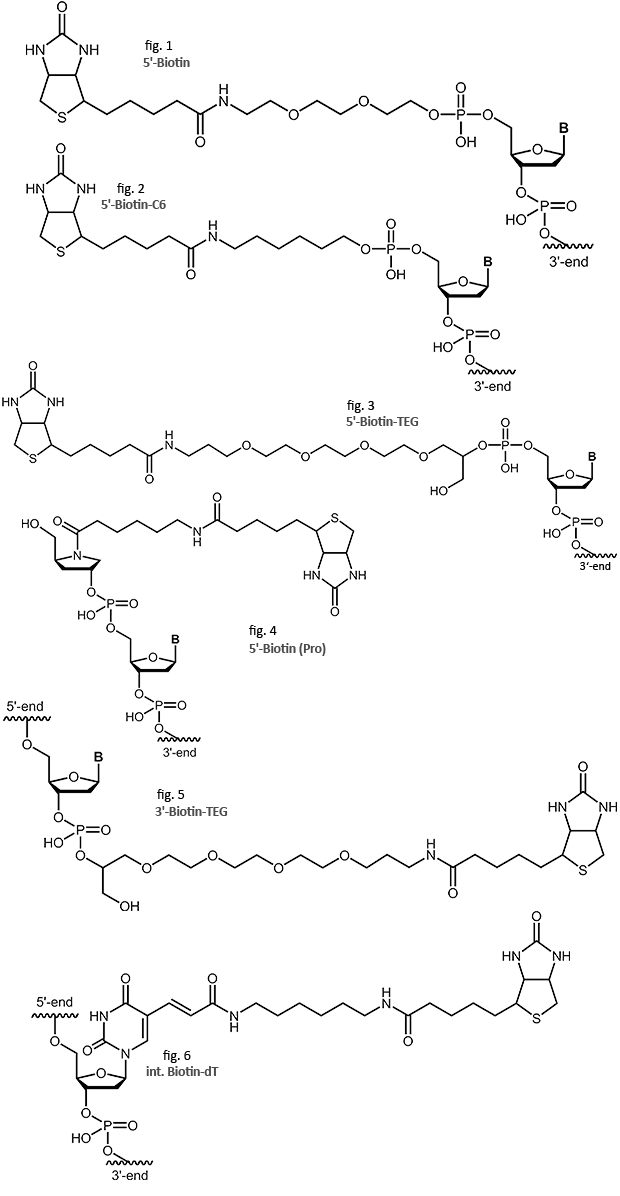
Oligonucleotides with Biotin modification
biomers.net offers different biotin modifications:
Biotin
The biotin-streptavidin binding is one of the strongest non-covalent interactions found in nature. The interaction has been explored for a wide variety of methods and applications in molecular biology, biochemistry and immunology. Biotinylated oligos are used as probes in hybridisation (Southern, Northern, ISH, or microarrays). Biotin-labeling also allows isolation, separation and purification of DNA or RNA with biotin/streptavidin affinity chromatography or magnetic beads. Specific methods like RACE or pyrosequencing use the capacity of biotin-based purification by binding the labeled DNA of interest to streptavidin coated surfaces.
Occurring naturally as Vitamin H (or B7), biotin can be linked to oligonucleotides in several ways. biomers.net normally uses the aminoethoxy-ethoxyethanol linker on the 5’ and the TEG linker on the 3’-end. But as the linker length and structure may influence the biotin/streptavidin interaction, in particular if surface-bound, Biotin-C6, Biotin-TEG and Biotin Pro are also available as alternatives.
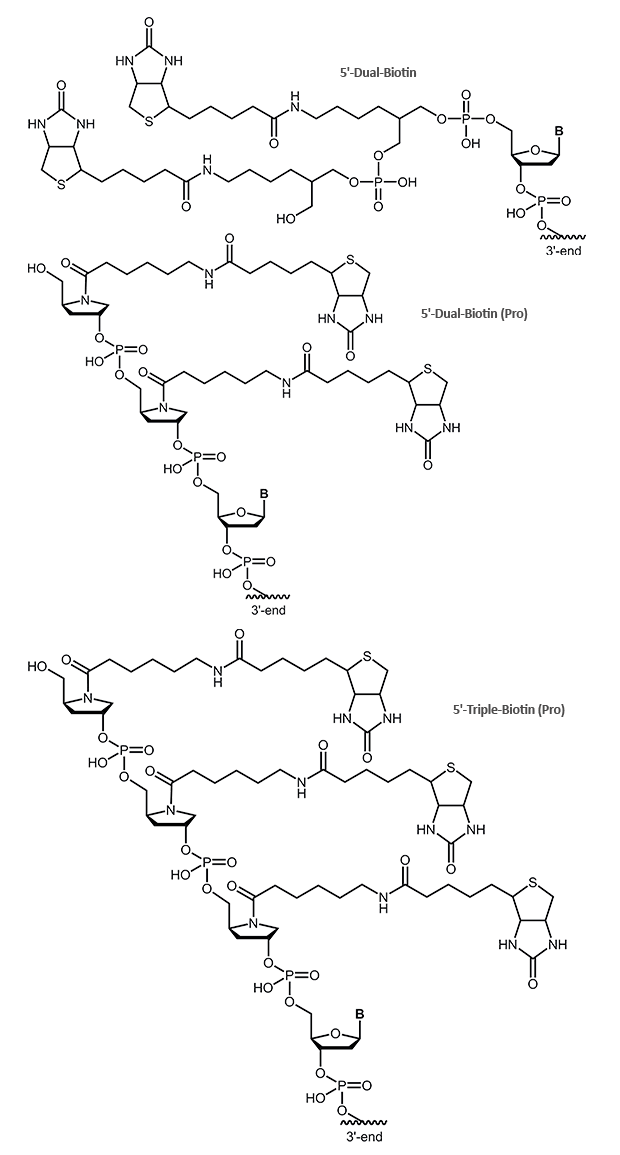
Dual-Biotin, Triple-Biotin
The dual-biotin modification adds two biotins to the 5’-end of the oligo. As both biotins can bind simultaneously and in a cooperative manner to two of the four binding sites on the streptavidin, an even stronger biotin/streptavidin interaction is achieved. Dual-biotin is used in methods that include a thermal denaturation step (e.g. PCR) with streptavidin-anchored probes or templates. It is commonly used in SAGE protocols (Serial Analysis of Gene Expression).
In the triple-biotin modification, three biotins are bound to the 5´-end of the oligonucleotide. Similar to the dual biotion, this biotin can also form a stronger bond to streptavidin.
Please note, the online ordering is not possible for the modifications 5´-Dual-Biotin (Pro) and 5´-Triple-Biotin (Pro). Please contact our customer support team for further information.
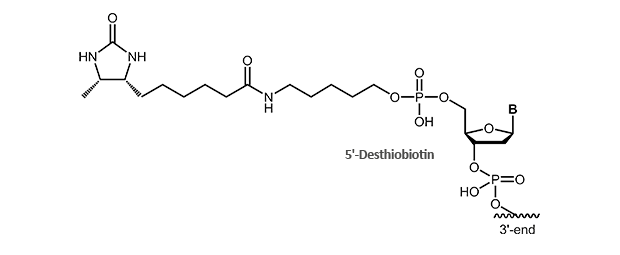
Desthiobiotin
Desthiobiotin lacks the S-atom in the biotin ring structure. It still binds specifically to streptavidin, but affinity is significantly decreased as compared to biotin. This allows purification of labeled nucleic acids and their hybrids in an exchange reaction with biotin using the biotin/streptavidin system. Being a very mild isolation procedure, recovery of DNA or RNA hybrids and sensitive conjugates is possible.
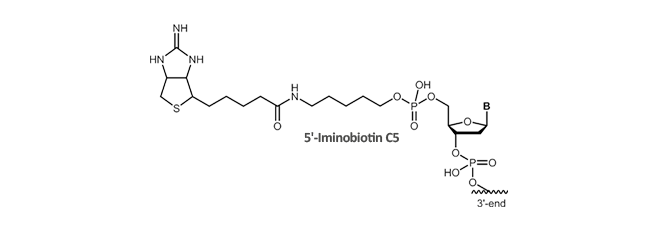
Iminobiotin
Iminobiotin is a cyclic guanidino analog of biotin and shows a high affinity specific binding to avidin or streptavidin at high pH (pH >9) and low affinity at low pH (pH <6). Due to the pH-dependent binding affinity, iminobiotin-modified proteins can be retained by avidin column at pH 9 to 11 and are eluted at pH 4 which represents a convenient purification method for these proteins under mild conditions preventing denaturation of purified proteins. At high pH, the free base form of iminobiotin retains the high-affinity binding properties of biotin whereas at low pH the salt form of iminobiotin only shows lower affinity to avidin enabling the break of the avidin-biotin binding.
Literature:
- Separation of complementary strands of plasmid DNA using the biotin-avidin system and its application to heteroduplex formation and RNA/DNA hybridizations in electron microscopy. Delius H, van Heerikhuizen H, Clarke J, Koller B; Nucleic Acids Res. (1985), 13, 5457-5469.
- In situ hybridization with biotinylated DNA probes: a rapid diagnostic test for adenovirus upper respiratory infections. Gomes SA, Nascimento JP, Siqueira MM, Krawczuk MM, Pereira HG, Russell WC; J. Virol. Methods (1985), 12, 105-110.
- Enhanced RACE method using specific enrichment by biotinylated oligonucleotides bound to streptavidin coated magnetic particles. Lankiewicz S, Gisselmann G, Hatt H; Nucleic Acids Res. (1997), 25, 2037-2038.
- Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B; Proc. Natl. Acad. Sci. USA. (2003), 100, 8817-8822.
- Direct labeling of RNA with multiple biotins allows sensitive expression profiling of acute leukemia class predictor genes. Cole K, Truong V, Barone D, McGall G; Nucleic Acids Res. (2004), 32, e86.
- Serial analysis of ribosomal sequence tags (SARST): a high-throughput method for profiling complex microbial communities.Neufeld JD, Yu Z, Lam W, Mohn WW; Environ. Microbiol. (2003), 6,131-144.
- Molecular interactions on microarrays. Southern EM, Mir K, Shchepinov MS; Nat. Genet. (1999), 21, 5-9.
- Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM, Haugland RP; Anal Biochem. (2002), 308, 343-57.
- Unbiased proteomic analysis of proteins interacting with the HIV-1 5′LTR sequence: role of the transcription factor Meis. Tacheny A, Michel S, Dieu M, Payen L, Arnould T, Renard P; Nucleic Acids Res. (2012), 40,1-18.
- Robust definition and identification of multiple oomycetes and fungi in environmental samples by using a novel cleavable padlock probe-based ligation detection assay. van Doom R, Slawiak M, Szemes M, Dullemans AM, Bonants P, Kowalchuk GA, Schoen CD; Appl. Environ. Microbiol. (2009), 75, 4185-4193.
- The Use of the 2-Iminobiotin-Avidin Interaction for the Selective Retrieval of Labeled Plasma Membrane Components. Orr GA; The Journal of biological chemistry (1981), Vol. 256, No. 2, Issue of January 25, pp. 761-766.
- Iminobiotin affinity columns and their application to retrieval of streptavidin. Hofmann K, Wood SW, Brinton CC, Montibeller JA, Finn FM; Proc Natl Acad Sci U S A (1980), 77(8): 4666–4668, doi: 10.1073/pnas.77.8.4666.
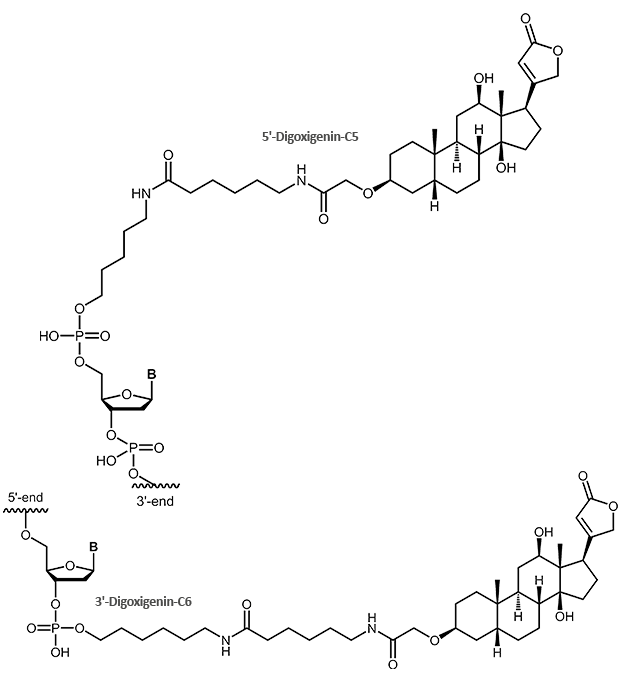
Digoxigenin (DIG)
Oligonucleotides with Digoxigenin modification
The steroid digoxigenin isolated from the plant Digitalis purpurea has been introduced as non-radioactive label for nucleic acids about 20 years ago. It got high acceptance and now is widely used to label DNA or RNA probes for mostly all hybridisation-based detection methods (Southern, Northern hybridisation, ISH or FISH).
 |
Literature:
- Digoxigenin as an alternative probe labeling for in situ hybridization. Komminoth P; Diagn Mol Pathol. (1992), 1, 142-150.
- Digoxigenin-end labeled oligonucleotides facilitate the detection of minimal residual disease in acute lymphoblastic leukemia. Nakao M, Janssen JWG, Schäfer M, Bartram CR; Leukemia (2000), 14, 767-772.
- Rapid chemiluminescent detection of mRNAs on northern blots with digoxigenin end-labelled oligonucleotides. Trayhurn P, Duncan JS, Nestor A, Thomas ME, Eastmond NC, Rayner DV; Electrophoresis (1995), 16, 341-344.
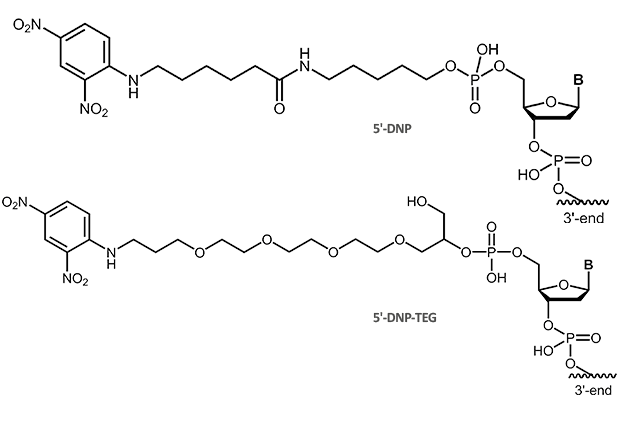
Dinitrophenol (DNP)
DNP
The small molecule dinitrophenol has been established for long as a label in immunohistochemistry and immunocytochemistry. It is also used to label oligonucleotide probes, in particular for the use in in-situ hybridisation (ISH or multicolor FISH).
 |
Literature:
- Synthesis and antibody-mediated detection of oligonucleotides containing multiple 2,4-dinitrophenyl reporter groups. Grzybowski J, Will DW, Randall RE, Smith CA, Brown T; Nucleic Acids Res. (1993), 21, 1705-1712.
- A comparative study of digoxigenin, 2,4-dinitrophenyl, and alkaline phosphatase deoxyoligonucleotide labels in non-radioisotopic in situ hybridization. Harper SJ, Bailey E, McKeen CM, Stewart AS, Pringle JH, Feehally J, Brown T, Bright R; J. Clin. Pathol. (1997), 5, 686-690.
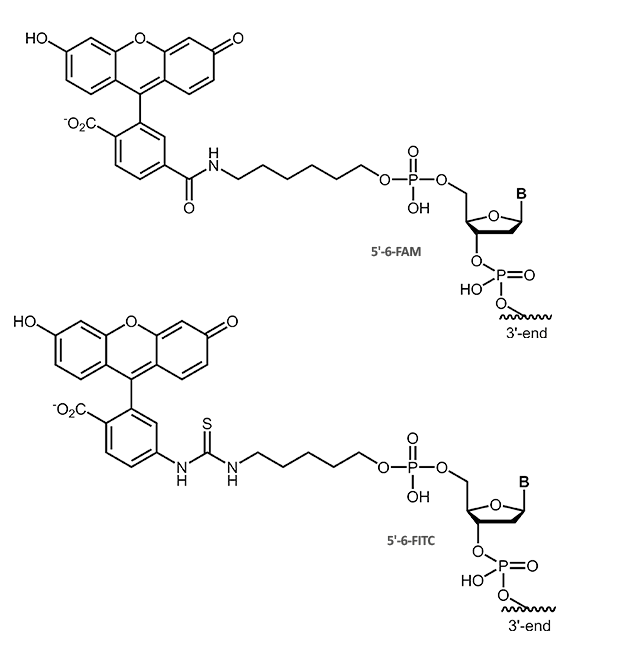
Fluorescein (Fam, FITC)
Fluorescein
The dye fluorescein can be used as label for direct fluorescence imaging. But it also serves as haptene-label that can be visualized with immuno-detection linked either to chemiluminiscence or colour formation.
The fluorescein label is available as 6-Fam or FITC, that differ only in the linkage of the fluorescein moiety to the oligonucleotide.
|
|
Literature:
- 3’-End fluorochromized and haptenized oligonucleotides as in situ hybridization probes for multiple simultaneous RNA detection. Dirks RW, Van Gijlswijk RPM, Vooijs MA, Smit AB, Bogerd J, Van Minnen J, Raap AK, Van der Ploeg M; Exp. Cell Res. (1991), 194, 310-315.
- Analysis of microRNA expression by in situ hybridization with RNA oligonucleotide probes. Thompson RC, Deo M, Turner DL; Methods (2007), 43, 153-161.
- In situ hybridization with fluoresceinated DNA. Wiegant, J, Ried T, Nederlof PM, Van der Ploeg M, Tanke HJ, Raap AK; Nucleic Acids Res. (1991), 19, 3237–3241.